Output Pickle File¶
On completion, the simulator outputs a pickle file of results for each gel lane (i.e. each individual experiment) to the results directory. Pickle is a format used by Python to save data. This page explains the format of the pickle data file.
You can load a pickle file by writing the following Python script:
import pickle
stackdata = pickle.load( open( "my_results_file.pkl", "rb" ) )
Each pickle file contains a dictionary with the following keys. Underneath each key is described the attribute that it maps to.
Dictionary keys to general properties of the simulation¶
stackdata[“bricks_add_order”]
List detailing the order in which strands were added to solution. E.g.
['s', 'p', 'X']
stackdata[“bricks_add_conc”]
List of Molar concentrations that strands above were added at. E.g.
[3e-07, 3e-07, 3e-07]
for 300nM concentrations.
stackdata[“bricks_wait_time”]
List of reaction wait times (seconds) that elapsed after each strand above was added, before the next washing step was carried out. E.g.
[1800, 1800, 1800]
for 30 minute wait times.
stackdata[“rule_name”]
List of reaction rule names in the reaction model describing the DNA stack chemistry.
['LH1', 'LH2', 'LH3', 'H1', 'H2', 'LH4', 'LH5', 'LH6', 'SD1', 'SD2', 'SD3', 'SD4', 'SD5', 'SD6', 'SD7', 'SD8', 'C1', 'C2', 'C3', 'C4']
Dictionary keys to chemistry state at each simulation stage¶
The remaining keys in the dictionary track the state of the model chemistry as each strand is added, one after the other. The keys have a number on the end, N, which indicates that the key refers to the chemistry state when N strands have been added after the original start strand.
The diagram below describes which chemistry state is referred to by a key followed by a number N:
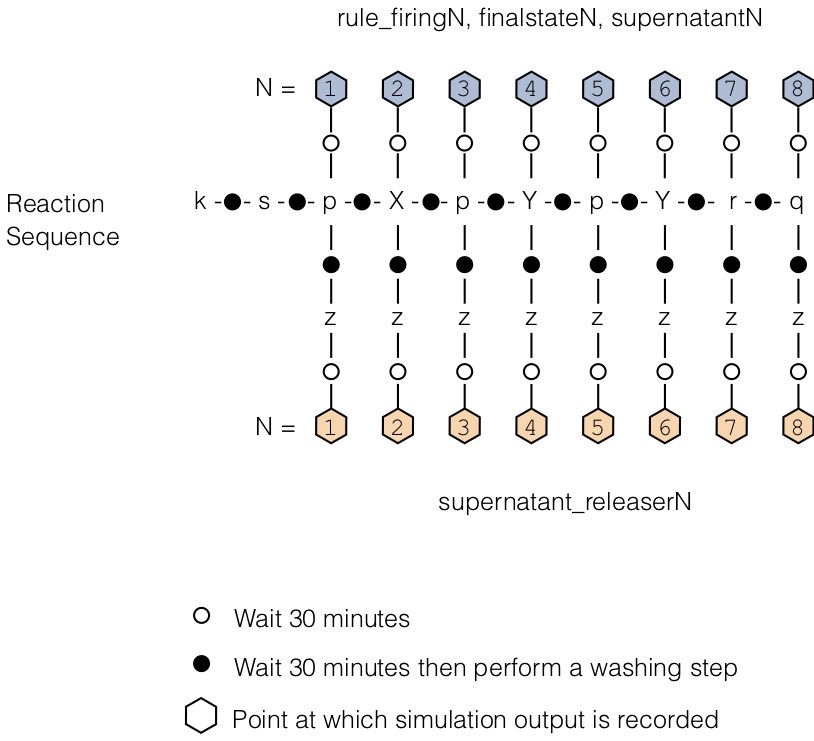
For example, keys rule_firing1, finalstate1, supernatant1, refer to the chemistry state after the first strand following start is added and a wait time elapses. Key supernatant_releaser1, on the other hand, refers to the chemistry state when the first strand following start is added and (i) a wait time elapses, (ii) a wash happens, (iii) releaser is added and (iv) a further wait time elapses.
Contents of the keys are described below.
stackdata[“rule_firingN”]
List of the total number of times each rule has fired, after the Nth strand following start is added and a waiting time elapses. E.g.
[234, 23, 1, 0, 12, 0, 34, 0, 345, 550, 0, 1, 0, 6, 0, 0, 12, 35, 0, 0]
stackdata[“finalstateN”]
Dictionary of the whole system state after the Nth strand following start has been added and a waiting time elapses. This is the state of every molecular species in the system, including all species tethered to beads and all species in supernatant.
The dictionary keys are species (strings) and the dictionary values are tuples giving (particle number, nM concentration). For example:
{'ks': (878, 9.719688673190916), 's': (187, 2.0701387037433956), 'p': (10634, 117.7211496021779), 'ksp': (13755, 152.27143245984172), 'sp': (2710, 30.000405813607497)}
stackdata[“supernatantN”]
Dictionary of species existing in the supernatant solution only, after the Nth strand following start has been added and a waiting time elapses. Format same as above. E.g.
{'s': (187, 2.0701387037433956), 'p': (10634, 117.7211496021779), 'sp': (2710, 30.000405813607497)}
The supernatant does not contain any stacks tethered to beads (species starting “k”).
stackdata[“supernatant_releaserN”]
Dictionary of species existing in the supernatant when, after the Nth strand following start has been added, the system is washed, releaser is added and then a waiting time elapses.
For example:
{'s': (501, 5.546200484360648), 'sp': (13363, 147.93189036429408), 'p': (2225, 24.63132949641206), 'z': (4897, 54.21106541300218)}
These species contain the stacks released from the beads, a fraction of any other supernatant species and any surplus releaser species (“z”).
The biotinylated DNA linker strands that are attached to sepharose beads (“k”) remain on the beads, possibly hybridised to releaser, and do not make it into the supernatant in this case.
Note
Kinetic trajectory information is NOT recorded in the results pickle file, as this tends to have an extremely large size. Only end states of each reaction stage are recorded.